The Molecular Identification of Pathogenic E. coli Isolated from Raw Cow Milk and Assessment Their Anti-susceptibility to Medical Plants at Al-Najaf city/ Iraq
Abstract
Background: Toxin-producing Shiga Escherichia coli has been identified as a new foodborne pathogen that poses a significant health risk to humans. Shiga toxin-producing Escherichia coli can be found in raw cow milk and its derivatives. A small number of Escherichia coli strains that produce shiga toxin are pathogenic.
Aim of study: The study aimed to see if there were any virulence genes in 50 milk samples that were typical of Entero-haemorrhagic E. coli and evaluate the Myrtus communis effects on these bacteria.
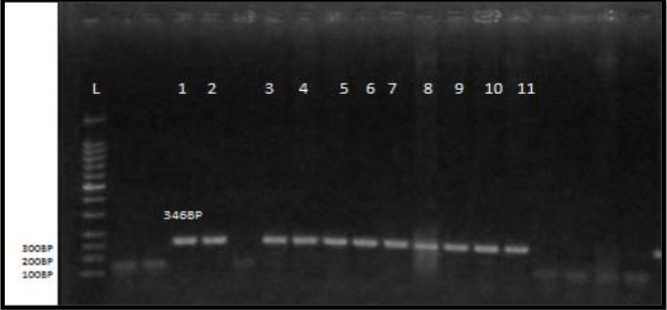
Materials and Method: Milk samples were used to isolate E. coli bacteria (n=27), biochemically analyzed, and genetically screened for virulence genes using a multiplex (PCR). The hydro-alcoholic extraction of Myrtus communis leaves was tested at four strengths, ranging from 20-50 mg/ml.
Results: The findings of the molecular profile indicated that (stx2) was found in 11 (40.7%), (hlyA) in 13 (48.2) and eae genes in 9 (33.3) of E. coli isolate, respectively. Treatment with an extract of this plant at a dosage of 50 mg/ml had the highest effect on Escherichia coli, which was significantly different from all other treatments.
Conclusions: The virulence genes shigatoxin-2(stx2), intimin(eae), and entero-hemolysin (hlyA) were found in Strains of E. coli isolated from milk, according to the findings of this study.